SAIGE-QTL Dynamic
SAIGE-QTL Dynamic is an advanced extension of SAIGE-QTL, which facilitates the analysis of heterogeneous genetic effects across different cell level and donor contexts directly from single-cell data efficiently, eliminating the need for pseudobulk aggregation.
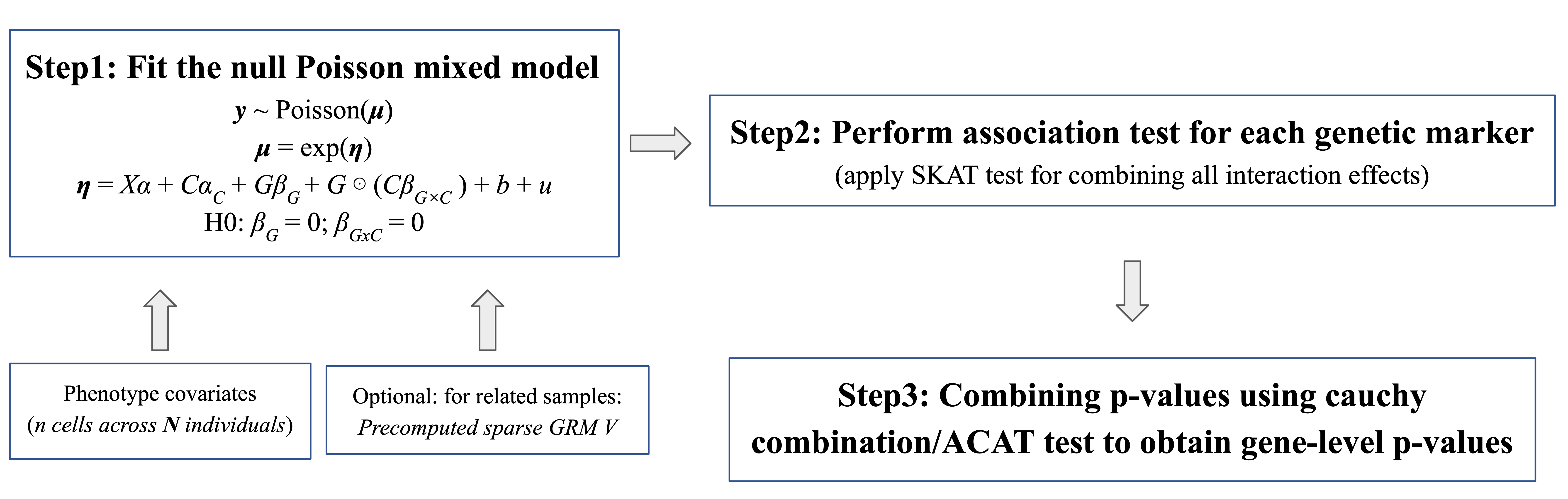
Analysis Workflow
Docker Installation
Prerequisites
- Docker installed on your system
- Access to pull images from Docker Hub
Pull the SAIGE-QTL Docker Image
The pre-built Docker image can be pulled directly from Docker Hub:
docker pull yijia0802/saigeqtldynamic0.2.5.1:latest
SAIGE-QTL Dynamic consists of three steps. Step 1 fits the null model, Step 2 obtains variant-level summary statistics for each gene, and Step 3 uses the ACAT method to calculate gene-level p-values. I’ve attached example shell scripts for running all three steps. Steps 1 and 2 can be executed directly within the Docker container.
step1_fitNULLGLMM_qtl.R
step2_tests_qtl.R
Step 3 can be excuted outside of the container with the provided script.
Support
This work is in progress. For any questions, please contact Christiana Liu: liuyijia@broadinstitute.org.